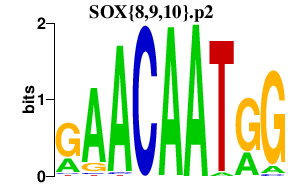
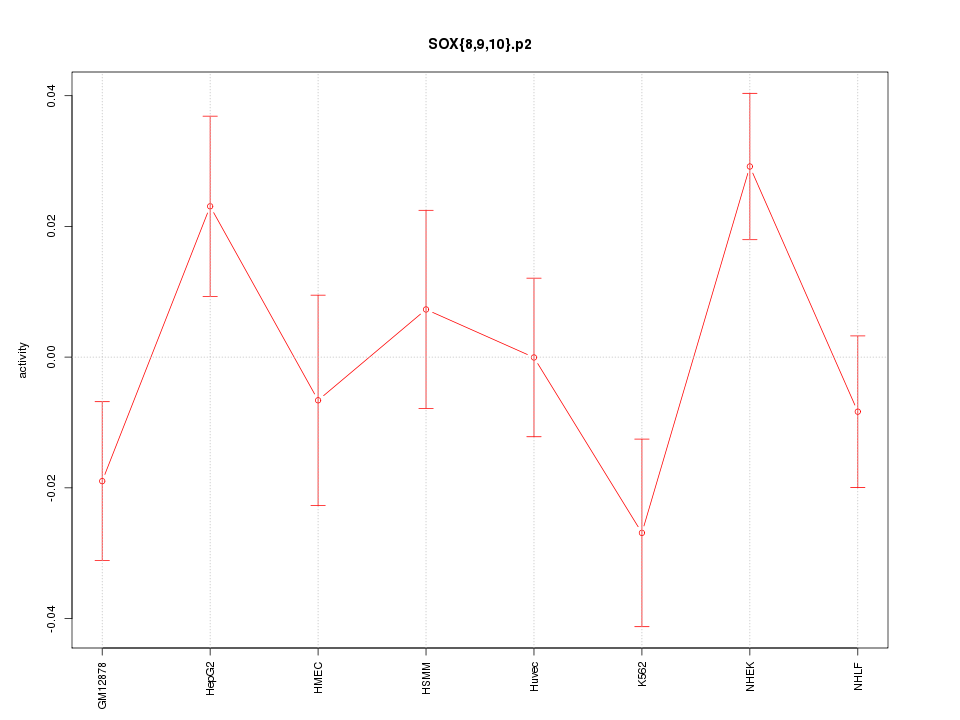
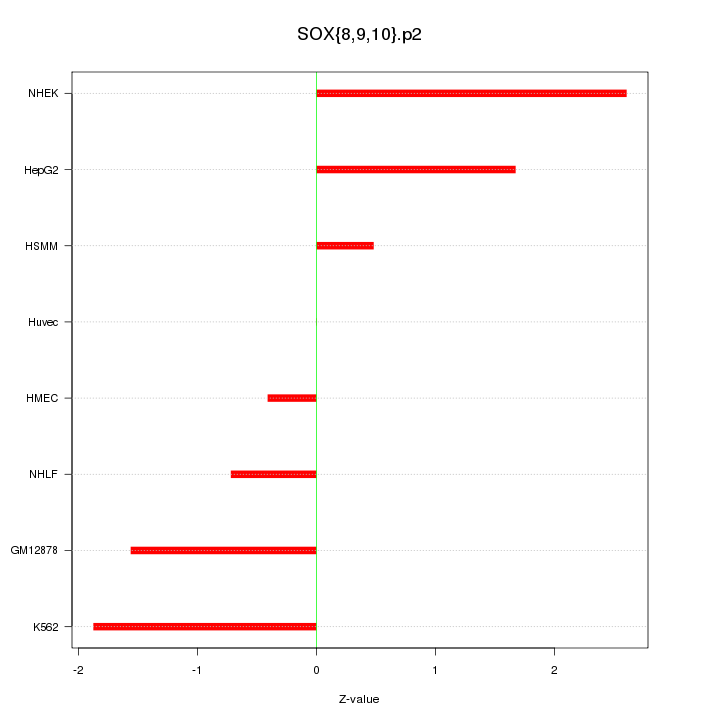
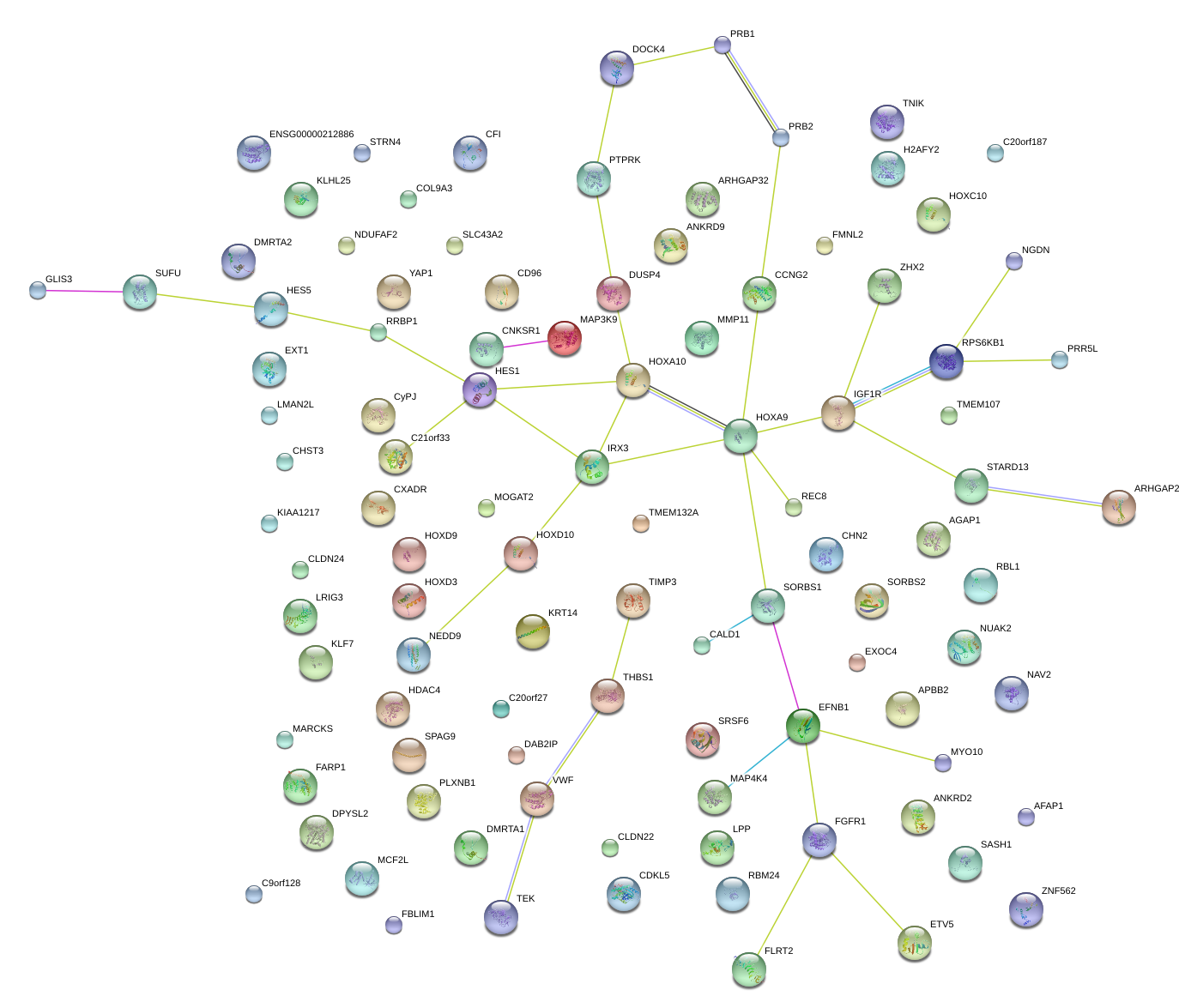
|
chr6_-_128883125
|
2.061
|
NM_002844
NM_001135648
|
PTPRK
|
protein tyrosine phosphatase, receptor type, K
|
|
chr9_+_123501186
|
1.387
|
|
DAB2IP
|
DAB2 interacting protein
|
|
chr2_-_239987557
|
1.240
|
NM_006037
|
HDAC4
|
histone deacetylase 4
|
|
chr2_+_101680594
|
1.193
|
NM_004834
NM_145686
NM_145687
|
MAP4K4
|
mitogen-activated protein kinase kinase kinase kinase 4
|
|
chr7_-_27171673
|
1.116
|
NM_152739
|
HOXA9
|
homeobox A9
|
|
chr11_+_20005704
|
1.079
|
|
NAV2
|
neuron navigator 2
|
|
chr3_-_187309442
|
1.039
|
NM_004454
|
ETV5
|
ets variant 5
|
|
chr10_-_97311084
|
1.035
|
NM_015385
NM_024991
|
SORBS1
|
sorbin and SH3 domain containing 1
|
|
chr7_-_27171648
|
1.024
|
|
HOXA9
|
homeobox A9
|
|
chr20_-_17610828
|
1.010
|
NM_001042576
NM_004587
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr7_-_27171600
|
0.998
|
|
HOXA9
|
homeobox A9
|
|
chr6_-_11340886
|
0.958
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr2_-_239987629
|
0.942
|
|
HDAC4
|
histone deacetylase 4
|
|
chr17_-_46479127
|
0.911
|
|
SPAG9
|
sperm associated antigen 9
|
|
chr4_-_186969041
|
0.901
|
NM_001145672
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr4_-_40631397
|
0.892
|
|
APBB2
|
amyloid beta (A4) precursor protein-binding, family B, member 2
|
|
chr12_+_52665196
|
0.874
|
NM_017409
|
HOXC10
|
homeobox C10
|
|
chr20_-_17610704
|
0.856
|
|
RRBP1
|
ribosome binding protein 1 homolog 180kDa (dog)
|
|
chr3_-_48445719
|
0.854
|
NM_002673
|
PLXNB1
|
plexin B1
|
|
chr6_-_11340869
|
0.852
|
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr11_+_101488380
|
0.847
|
NM_001195045
|
YAP1
|
Yes-associated protein 1
|
|
chr2_+_152900076
|
0.838
|
|
FMNL2
|
formin-like 2
|
|
chr14_-_89490780
|
0.826
|
NM_145231
|
EFCAB11
|
EF-hand calcium binding domain 11
|
|
chr2_+_176737050
|
0.810
|
NM_006898
|
HOXD3
|
homeobox D3
|
|
chr20_+_60918836
|
0.802
|
NM_001853
|
COL9A3
|
collagen, type IX, alpha 3
|
|
chr22_+_22445035
|
0.790
|
NM_005940
|
MMP11
|
matrix metallopeptidase 11 (stromelysin 3)
|
|
chr14_+_23711044
|
0.778
|
NM_001048205
NM_005132
|
REC8
|
REC8 homolog (yeast)
|
|
chr14_+_23008723
|
0.776
|
NM_001042635
NM_015514
|
NGDN
|
neuroguidin, EIF4E binding protein
|
|
chr6_-_11340887
|
0.764
|
NM_006403
NM_182966
|
NEDD9
|
neural precursor cell expressed, developmentally down-regulated 9
|
|
chr5_-_16989371
|
0.759
|
NM_012334
|
MYO10
|
myosin X
|
|
chr4_-_7991992
|
0.758
|
NM_001134647
NM_198595
|
AFAP1
|
actin filament associated protein 1
|
|
chr22_+_31527678
|
0.746
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr7_+_134114912
|
0.744
|
|
CALD1
|
caldesmon 1
|
|
chr1_-_203557379
|
0.743
|
NM_030952
|
NUAK2
|
NUAK family, SNF1-like kinase, 2
|
|
chr13_+_97593469
|
0.740
|
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr9_+_27099138
|
0.733
|
NM_000459
|
TEK
|
TEK tyrosine kinase, endothelial
|
|
chr8_-_29262240
|
0.728
|
NM_057158
|
DUSP4
|
dual specificity phosphatase 4
|
|
chr21_+_17807167
|
0.727
|
NM_001338
|
CXADR
|
coxsackie virus and adenovirus receptor
|
|
chr17_-_36996611
|
0.726
|
NM_000526
|
KRT14
|
keratin 14
|
|
chrX_+_67965517
|
0.719
|
NM_004429
|
EFNB1
|
ephrin-B1
|
|
chr3_+_189425886
|
0.714
|
NM_001167671
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr17_-_1478293
|
0.706
|
|
SLC43A2
|
solute carrier family 43, member 2
|
|
chr7_+_29486010
|
0.698
|
NM_001039936
|
CHN2
|
chimerin (chimaerin) 2
|
|
chr7_-_111633428
|
0.698
|
|
DOCK4
|
dedicator of cytokinesis 4
|
|
chr13_+_112704077
|
0.697
|
|
MCF2L
|
MCF.2 cell line derived transforming sequence-like
|
|
chr12_-_57600564
|
0.694
|
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr4_-_186969202
|
0.693
|
NM_001145671
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr10_+_104253716
|
0.683
|
|
SUFU
|
suppressor of fused homolog (Drosophila)
|
|
chr3_+_189354167
|
0.682
|
NM_001167672
|
LPP
|
LIM domain containing preferred translocation partner in lipoma
|
|
chr12_-_11399776
|
0.676
|
NM_005039
NM_199353
NM_199354
|
PRB1
|
proline-rich protein BstNI subfamily 1
|
|
chrX_+_18353623
|
0.673
|
NM_003159
|
CDKL5
|
cyclin-dependent kinase-like 5
|
|
chr14_-_70345485
|
0.672
|
NM_033141
|
MAP3K9
|
mitogen-activated protein kinase kinase kinase 9
|
|
chr11_-_128567302
|
0.671
|
NM_001142685
|
ARHGAP32
|
Rho GTPase activating protein 32
|
|
chr2_-_207739815
|
0.664
|
|
KLF7
|
Kruppel-like factor 7 (ubiquitous)
|
|
chr10_+_71482542
|
0.660
|
|
H2AFY2
|
H2A histone family, member Y2
|
|
chr8_+_26491317
|
0.655
|
NM_001386
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr10_+_73393998
|
0.650
|
NM_004273
|
CHST3
|
carbohydrate (chondroitin 6) sulfotransferase 3
|
|
chr16_-_49205063
|
0.648
|
|
|
|
|
chr1_+_15957832
|
0.645
|
NM_017556
|
FBLIM1
|
filamin binding LIM protein 1
|
|
chr8_+_26491383
|
0.631
|
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr6_+_148705421
|
0.630
|
NM_015278
|
SASH1
|
SAM and SH3 domain containing 1
|
|
chr5_-_50714836
|
0.629
|
|
LOC642366
|
hypothetical LOC642366
|
|
chr8_-_38444982
|
0.629
|
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr5_+_60276695
|
0.622
|
NM_174889
|
NDUFAF2
|
NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 2
|
|
chr16_-_52877868
|
0.618
|
NM_024336
|
IRX3
|
iroquois homeobox 3
|
|
chr6_+_114285209
|
0.617
|
NM_002356
|
MARCKS
|
myristoylated alanine-rich protein kinase C substrate
|
|
chr4_-_184478920
|
0.616
|
NM_001111319
|
CLDN22
|
claudin 22
|
|
chr22_+_31527744
|
0.615
|
|
TIMP3
|
TIMP metallopeptidase inhibitor 3
|
|
chr2_-_96769502
|
0.613
|
|
LMAN2L
|
lectin, mannose-binding 2-like
|
|
chr1_-_2451543
|
0.611
|
NM_001010926
|
HES5
|
hairy and enhancer of split 5 (Drosophila)
|
|
chr13_-_32658215
|
0.610
|
NM_178007
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr3_+_195336625
|
0.610
|
NM_005524
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr14_+_85066265
|
0.608
|
|
FLRT2
|
fibronectin leucine rich transmembrane protein 2
|
|
chr1_-_94475504
|
0.608
|
|
ARHGAP29
|
Rho GTPase activating protein 29
|
|
chr3_+_112743729
|
0.608
|
|
CD96
|
CD96 molecule
|
|
chr9_-_35818728
|
0.604
|
NM_001012446
|
C9orf128
|
chromosome 9 open reading frame 128
|
|
chr8_-_38445272
|
0.602
|
|
FGFR1
|
fibroblast growth factor receptor 1
|
|
chr10_+_71482362
|
0.601
|
NM_018649
|
H2AFY2
|
H2A histone family, member Y2
|
|
chr1_-_50661716
|
0.593
|
NM_032110
|
DMRTA2
|
DMRT-like family A2
|
|
chr2_+_236068029
|
0.583
|
|
AGAP1
|
ArfGAP with GTPase domain, ankyrin repeat and PH domain 1
|
|
chr11_+_60448487
|
0.581
|
NM_017870
NM_178031
|
TMEM132A
|
transmembrane protein 132A
|
|
chr11_+_36274300
|
0.581
|
NM_001160167
|
PRR5L
|
proline rich 5 like
|
|
chr10_+_24538094
|
0.580
|
|
KIAA1217
|
KIAA1217
|
|
chr10_+_24537725
|
0.578
|
NM_001098501
NM_019590
|
KIAA1217
|
KIAA1217
|
|
chr14_-_102045849
|
0.577
|
NM_152326
|
ANKRD9
|
ankyrin repeat domain 9
|
|
chr1_+_26376560
|
0.576
|
NM_006314
|
CNKSR1
|
connector enhancer of kinase suppressor of Ras 1
|
|
chr12_-_6103945
|
0.576
|
NM_000552
|
VWF
|
von Willebrand factor
|
|
chr6_+_17390910
|
0.576
|
NM_153020
|
RBM24
|
RNA binding motif protein 24
|
|
chr2_-_96769526
|
0.576
|
NM_001142292
NM_030805
|
LMAN2L
|
lectin, mannose-binding 2-like
|
|
chr8_+_124032830
|
0.575
|
|
ZHX2
|
zinc fingers and homeoboxes 2
|
|
chr8_-_119192985
|
0.570
|
NM_000127
|
EXT1
|
exostosin 1
|
|
chr15_+_37660568
|
0.569
|
NM_003246
|
THBS1
|
thrombospondin 1
|
|
chr10_+_104253706
|
0.562
|
NM_001178133
NM_016169
|
SUFU
|
suppressor of fused homolog (Drosophila)
|
|
chr15_-_84139182
|
0.562
|
NM_022480
|
KLHL25
|
kelch-like 25 (Drosophila)
|
|
chr7_-_27186400
|
0.562
|
NM_153715
|
HOXA10
|
homeobox A10
|
|
chr15_+_97009194
|
0.559
|
|
IGF1R
|
insulin-like growth factor 1 receptor
|
|
chr15_-_84139021
|
0.556
|
|
KLHL25
|
kelch-like 25 (Drosophila)
|
|
chr20_-_3696344
|
0.556
|
NM_001039140
|
C20orf27
|
chromosome 20 open reading frame 27
|
|
chr4_-_110942576
|
0.555
|
|
CFI
|
complement factor I
|
|
chr9_-_4142182
|
0.552
|
NM_152629
|
GLIS3
|
GLIS family zinc finger 3
|
|
chr16_-_52877829
|
0.548
|
|
IRX3
|
iroquois homeobox 3
|
|
chr4_+_78298326
|
0.545
|
|
CCNG2
|
cyclin G2
|
|
chr20_+_41519947
|
0.544
|
|
SRSF6
|
serine/arginine-rich splicing factor 6
|
|
chr11_+_75106512
|
0.543
|
NM_025098
|
MOGAT2
|
monoacylglycerol O-acyltransferase 2
|
|
chr2_-_164300758
|
0.540
|
NM_018086
|
FIGN
|
fidgetin
|
|
chr2_-_21106291
|
0.536
|
|
APOB
|
apolipoprotein B (including Ag(x) antigen)
|
|
chr12_+_62524778
|
0.535
|
NM_020762
|
SRGAP1
|
SLIT-ROBO Rho GTPase activating protein 1
|
|
chr3_-_150422474
|
0.534
|
NM_000096
|
CP
|
ceruloplasmin (ferroxidase)
|
|
chr14_-_102045749
|
0.532
|
|
ANKRD9
|
ankyrin repeat domain 9
|
|
chr7_+_148613304
|
0.530
|
|
LOC155060
|
AI894139 pseudogene
|
|
chr4_-_170314382
|
0.525
|
|
SH3RF1
|
SH3 domain containing ring finger 1
|
|
chr7_+_134114957
|
0.523
|
|
CALD1
|
caldesmon 1
|
|
chr1_-_202730135
|
0.518
|
|
PIK3C2B
|
phosphoinositide-3-kinase, class 2, beta polypeptide
|
|
chr2_-_215953818
|
0.517
|
|
FN1
|
fibronectin 1
|
|
chr9_+_115303527
|
0.517
|
NM_130795
|
RGS3
|
regulator of G-protein signaling 3
|
|
chr17_+_18069593
|
0.516
|
NM_004140
|
LLGL1
|
lethal giant larvae homolog 1 (Drosophila)
|
|
chr14_-_23090606
|
0.513
|
NM_033400
|
ZFHX2
|
zinc finger homeobox 2
|
|
chr15_+_83724743
|
0.511
|
NM_006738
NM_007200
|
AKAP13
|
A kinase (PRKA) anchor protein 13
|
|
chr10_+_24795446
|
0.509
|
|
KIAA1217
|
KIAA1217
|
|
chr5_+_50714996
|
0.509
|
|
ISL1
|
ISL LIM homeobox 1
|
|
chr17_+_39778191
|
0.506
|
|
GRN
|
granulin
|
|
chr14_-_93859394
|
0.505
|
NM_001756
|
SERPINA6
|
serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 6
|
|
chr1_-_95165089
|
0.505
|
NM_001839
|
CNN3
|
calponin 3, acidic
|
|
chr6_+_17390273
|
0.502
|
NM_001143941
|
RBM24
|
RNA binding motif protein 24
|
|
chr3_-_124821868
|
0.501
|
|
MYLK
|
myosin light chain kinase
|
|
chr17_-_1478859
|
0.500
|
NM_152346
|
SLC43A2
|
solute carrier family 43, member 2
|
|
chr3_-_151171576
|
0.497
|
|
PFN2
|
profilin 2
|
|
chr8_+_26491507
|
0.495
|
|
DPYSL2
|
dihydropyrimidinase-like 2
|
|
chr20_-_13919256
|
0.494
|
NM_025229
|
SEL1L2
|
sel-1 suppressor of lin-12-like 2 (C. elegans)
|
|
chr4_-_110942783
|
0.490
|
NM_000204
|
CFI
|
complement factor I
|
|
chr14_+_23008742
|
0.489
|
|
NGDN
|
neuroguidin, EIF4E binding protein
|
|
chr19_-_5291700
|
0.488
|
NM_002850
NM_130853
NM_130854
NM_130855
|
PTPRS
|
protein tyrosine phosphatase, receptor type, S
|
|
chr11_-_27450808
|
0.486
|
NM_018490
|
LGR4
|
leucine-rich repeat containing G protein-coupled receptor 4
|
|
chr12_-_10142779
|
0.484
|
NM_016511
|
CLEC1A
|
C-type lectin domain family 1, member A
|
|
chr22_-_41672990
|
0.480
|
NM_007229
|
PACSIN2
|
protein kinase C and casein kinase substrate in neurons 2
|
|
chr13_+_97593711
|
0.479
|
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr17_-_10393664
|
0.478
|
NM_001100112
NM_017534
|
MYH2
|
myosin, heavy chain 2, skeletal muscle, adult
|
|
chr9_+_80101810
|
0.476
|
NM_021154
NM_058179
|
PSAT1
|
phosphoserine aminotransferase 1
|
|
chr6_-_80001116
|
0.471
|
NM_004242
NM_138730
|
HMGN3
|
high mobility group nucleosomal binding domain 3
|
|
chr9_+_136358145
|
0.469
|
|
RXRA
|
retinoid X receptor, alpha
|
|
chr13_-_32678142
|
0.467
|
NM_052851
|
STARD13
|
StAR-related lipid transfer (START) domain containing 13
|
|
chr11_-_74914514
|
0.466
|
|
GDPD5
|
glycerophosphodiester phosphodiesterase domain containing 5
|
|
chr15_-_68781662
|
0.465
|
NM_001008224
|
UACA
|
uveal autoantigen with coiled-coil domains and ankyrin repeats
|
|
chr12_-_11439752
|
0.461
|
NM_006248
|
PRB1
PRB2
|
proline-rich protein BstNI subfamily 1
proline-rich protein BstNI subfamily 2
|
|
chr14_-_92284626
|
0.460
|
NM_001008530
NM_005606
|
LGMN
|
legumain
|
|
chr18_+_7936885
|
0.456
|
|
PTPRM
|
protein tyrosine phosphatase, receptor type, M
|
|
chr1_-_95165037
|
0.451
|
|
CNN3
|
calponin 3, acidic
|
|
chr11_+_4617520
|
0.451
|
NM_001004751
|
OR51D1
|
olfactory receptor, family 51, subfamily D, member 1
|
|
chr10_+_24795465
|
0.447
|
|
KIAA1217
|
KIAA1217
|
|
chr17_+_39778180
|
0.447
|
|
GRN
|
granulin
|
|
chr2_-_239988269
|
0.443
|
|
HDAC4
|
histone deacetylase 4
|
|
chr5_+_50714714
|
0.442
|
NM_002202
|
ISL1
|
ISL LIM homeobox 1
|
|
chr1_+_33494745
|
0.441
|
NM_152493
|
ZNF362
|
zinc finger protein 362
|
|
chr7_+_96473225
|
0.440
|
NM_005222
|
DLX6
|
distal-less homeobox 6
|
|
chr1_-_85703198
|
0.439
|
|
DDAH1
|
dimethylarginine dimethylaminohydrolase 1
|
|
chr13_-_40138607
|
0.437
|
NM_002015
|
FOXO1
|
forkhead box O1
|
|
chr12_-_57600336
|
0.436
|
NM_153377
|
LRIG3
|
leucine-rich repeats and immunoglobulin-like domains 3
|
|
chr5_-_125958503
|
0.433
|
NM_001182
|
ALDH7A1
|
aldehyde dehydrogenase 7 family, member A1
|
|
chr14_+_23710924
|
0.427
|
|
REC8
|
REC8 homolog (yeast)
|
|
chr3_-_71625318
|
0.426
|
|
FOXP1
|
forkhead box P1
|
|
chr3_+_195336631
|
0.425
|
|
HES1
|
hairy and enhancer of split 1, (Drosophila)
|
|
chr4_-_5941043
|
0.424
|
|
CRMP1
|
collapsin response mediator protein 1
|
|
chr6_-_110843432
|
0.423
|
NM_003649
NM_004032
|
DDO
|
D-aspartate oxidase
|
|
chr17_-_71017509
|
0.422
|
NM_001142643
|
CASKIN2
|
CASK interacting protein 2
|
|
chr2_-_1727220
|
0.422
|
|
PXDN
|
peroxidasin homolog (Drosophila)
|
|
chr19_-_63205948
|
0.419
|
|
ZNF606
|
zinc finger protein 606
|
|
chr2_+_228386800
|
0.404
|
NM_001130046
NM_004591
|
CCL20
|
chemokine (C-C motif) ligand 20
|
|
chr17_-_59818033
|
0.403
|
|
PECAM1
|
platelet/endothelial cell adhesion molecule
|
|
chr19_-_10352212
|
0.402
|
NM_003331
|
TYK2
|
tyrosine kinase 2
|
|
chr18_-_72857665
|
0.402
|
|
MBP
|
myelin basic protein
|
|
chr7_-_41709191
|
0.397
|
NM_002192
|
INHBA
|
inhibin, beta A
|
|
chr10_-_105835525
|
0.397
|
|
COL17A1
|
collagen, type XVII, alpha 1
|
|
chr17_-_37196463
|
0.395
|
NM_002230
NM_021991
|
JUP
|
junction plakoglobin
|
|
chr11_-_116213511
|
0.390
|
NM_000039
|
APOA1
|
apolipoprotein A-I
|
|
chr17_+_39778153
|
0.390
|
|
GRN
|
granulin
|
|
chr19_-_10352172
|
0.389
|
|
TYK2
|
tyrosine kinase 2
|
|
chr10_-_103864651
|
0.389
|
NM_003893
|
LDB1
|
LIM domain binding 1
|
|
chr22_+_36422940
|
0.388
|
NM_001039141
|
TRIOBP
|
TRIO and F-actin binding protein
|
|
chr19_+_7566698
|
0.384
|
NM_001080429
NM_020902
|
KIAA1543
|
KIAA1543
|
|
chr1_+_176577228
|
0.383
|
NM_004841
|
RASAL2
|
RAS protein activator like 2
|
|
chr2_+_219532590
|
0.383
|
NM_003936
|
CDK5R2
|
cyclin-dependent kinase 5, regulatory subunit 2 (p39)
|
|
chr2_+_74734861
|
0.382
|
NM_004263
|
SEMA4F
|
sema domain, immunoglobulin domain (Ig), transmembrane domain (TM) and short cytoplasmic domain, (semaphorin) 4F
|
|
chr13_+_28900776
|
0.379
|
NM_015233
|
MTUS2
|
microtubule associated tumor suppressor candidate 2
|
|
chr22_-_29052863
|
0.379
|
NM_031937
|
TBC1D10A
|
TBC1 domain family, member 10A
|
|
chr13_+_97593411
|
0.373
|
NM_001001715
NM_005766
|
FARP1
|
FERM, RhoGEF (ARHGEF) and pleckstrin domain protein 1 (chondrocyte-derived)
|
|
chr2_-_204108151
|
0.373
|
NM_203365
NM_213589
|
RAPH1
|
Ras association (RalGDS/AF-6) and pleckstrin homology domains 1
|
|
chr4_-_186815116
|
0.373
|
NM_001145674
NM_001145675
|
SORBS2
|
sorbin and SH3 domain containing 2
|
|
chr16_-_70168449
|
0.373
|
NM_000353
|
TAT
|
tyrosine aminotransferase
|
|
chr9_-_73573121
|
0.371
|
NM_001135820
NM_013390
|
TMEM2
|
transmembrane protein 2
|
|
chr15_-_53822468
|
0.370
|
NM_173814
|
PRTG
|
protogenin
|
|
chr5_-_147142249
|
0.367
|
|
JAKMIP2
|
janus kinase and microtubule interacting protein 2
|
|
chr8_+_70541412
|
0.367
|
NM_001128204
NM_015170
|
SULF1
|
sulfatase 1
|
|
chr6_+_37895244
|
0.364
|
NM_021943
|
ZFAND3
|
zinc finger, AN1-type domain 3
|
|
chr16_+_464844
|
0.360
|
NM_001142272
|
RAB11FIP3
|
RAB11 family interacting protein 3 (class II)
|
|
chr10_+_5126589
|
0.360
|
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr16_-_30705707
|
0.360
|
NM_001080417
|
ZNF629
|
zinc finger protein 629
|
|
chr8_+_70541631
|
0.358
|
|
SULF1
|
sulfatase 1
|
|
chr2_-_175337386
|
0.356
|
NM_000079
NM_001039523
|
CHRNA1
|
cholinergic receptor, nicotinic, alpha 1 (muscle)
|
|
chr10_+_5126613
|
0.354
|
|
AKR1C3
|
aldo-keto reductase family 1, member C3 (3-alpha hydroxysteroid dehydrogenase, type II)
|
|
chr17_+_37373195
|
0.353
|
|
|
|